── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
✔ ggplot2 3.4.0 ✔ purrr 1.0.1
✔ tibble 3.2.1 ✔ dplyr 1.1.1
✔ tidyr 1.2.1 ✔ stringr 1.5.0
✔ readr 2.1.3 ✔ forcats 0.5.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
Attaching package: 'lubridate'
The following objects are masked from 'package:base':
date, intersect, setdiff, union
This code: creates a dataframe of dispensaries from 2013 - 2023
- retrieves all of the filenames from the working directory
- retains the filename as new variable
- combines the listed excel files from a defined directory into a single dataframe
- creates a new date variable extracted from the filename
- merges like variables with different column names together and overrides the N/As
- cleans up the business names, and classifies the observation drawn from the license number prefix
- finds missing business names and replaces the null values with features extracted from similar observations
- finds the missing zips codes and replaces the null values with matching license numbers
This section of my methodology was the most time intensive. Having quality historical data is the foundation of my project. Calculating the number of dispensaries is a key variable for the linear regression model.
NIMBY’s Method:
The data on dispensary locations come from the Colorado Department of Revenue, which starting in January 2013 has published a monthly list of each active medical and recreational license. Overall, our sample covers 48 months between 2013 and 2016 for 143 census tracts in the City of Denver.
[@brinkman2017]
NIMBY’s data were accessed at the no longer active Colorado.gov page: in January 2017.
My Method:
Data Source: Colorado Department of Revenue: Medical and Retail Marijuana Licencees
Accessed April 2023
Before running the code below, I downloaded the dispensary files from the Colorado Department of Revenue’s monthly list of med licensee data archive, and manually adjusted the naming convention to reflect the file’s time period.
install packages:
combine files into a single dataframe, with a new date field:
adapted from “Pass chunk of filename to column name in a loop”
Code
# set working directory for this chunk only
setwd("~/gvsu/winter 23/CIS 635/NIMBY/cleaning/dsp_data")
dsp <- dir_ls() %>% set_names() # grab all of the file names
# the .id parameter creates a new column called "filename"
all_dsps <- map_df(dsp, ~read_excel(.x), .id = "filename")New names:
New names:
New names:
• `` -> `...8`Code
# extract the date from the file name and create a new date column
all_dsps <- all_dsps %>%
mutate(date = str_extract(filename, "\\d{6}"),
date = as.Date(paste0("01", date), format = "%d%m%Y"))
create uniform observation fields:
The ledgers vary throughout the years. In order to create a cohesive database, I needed to ensure like fields held the same naming conventions so they can be aggregated together down the road.
| dispensary record fields: | |
|---|---|
| city | dba (doing business as) |
| licensee | license number |
| zip code |
adapted from “How to Use the coalesce() Function in dplyr”
Code
# use coalesce to combine like columns and override the N/A
all_dsps<- all_dsps %>%
mutate(dba = coalesce(DBA,`DOING BUSINESS AS`)) %>%
mutate(city = coalesce(CITY, City)) %>%
mutate(licensee = coalesce(Licensee, Licensees, LICENSEE, `Licensee Name` )) %>%
mutate(zip = coalesce(ZIP, Zip, ZipCode)) %>%
mutate(license_no = coalesce(`Lic #`,`LICENSE #`, `License Number`,LicenseNumber, license_no, `License #` ))
#make a year column / variable
all_dsps$year <- substr(all_dsps$date, 1,4)
# write.csv(all_dsps, "firstbatch.csv")
# select the relevant fields
all_dsps <- all_dsps %>%
select(1, 26, 37:42)
create uniform business names
Another inconsistent feature of the ledger includes many variations in the spelling of a business name. Throughout this project, I became vary familiar with the cannabis related business names and puns. I assume that if a similarly spelled business holds the same licence number, I can override that name to reflect the predominant naming convention. This tedious task was crucial in allowing me to match businesses that sell both medical and recreational products under different licenses.
Code
# same case
all_dsps$dba <- toupper(all_dsps$dba) # DBA = "Doing Business As"
# replace chaotic naming conventions
all_dsps$dba <- gsub("3D CENTER","3D CANNABIS CENTER", all_dsps$dba)
all_dsps$dba <- gsub("3D CANNABIS CENTER AND 3D DISPENSARY","3D CANNABIS CENTER", all_dsps$dba)
all_dsps$dba <- gsub("3D DISPENSARY AND/OR 3D CANNABIS CENTER","3D CANNABIS CENTER", all_dsps$dba)
all_dsps$dba <- gsub("21+","", all_dsps$dba)
all_dsps$dba <- gsub(" LLC","", all_dsps$dba)
# only remove LLC if nothing else follows
all_dsps$dba <- gsub("LLC(?!.)$", "", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("&","AND", all_dsps$dba)
all_dsps$dba <- gsub(",","", all_dsps$dba)
all_dsps$dba <- gsub(" INC","", all_dsps$dba)
all_dsps$dba <- gsub("INC","", all_dsps$dba)
all_dsps$dba <- gsub("'","", all_dsps$dba)
all_dsps$dba <- gsub("?S","S", all_dsps$dba)
all_dsps$dba <- gsub("-","", all_dsps$dba)
all_dsps$dba <- gsub(":","", all_dsps$dba)
all_dsps$dba <- gsub("COMPANY","", all_dsps$dba)
all_dsps$dba <- gsub(" CO(?!.)$", "", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("CO(?!.)$", "", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("CENTERS","CENTER", all_dsps$dba)
all_dsps$dba <- gsub("LLP(?!.)$", "", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("B.GOOD","BGOOD", all_dsps$dba)
all_dsps$dba <- gsub("B.?GOOD","BGOOD", all_dsps$dba)
all_dsps$dba <- gsub("B GOOD","BGOOD", all_dsps$dba)
all_dsps$dba <- gsub("DEN REC","DENREC", all_dsps$dba)
all_dsps$dba <- gsub("111","1:11", all_dsps$dba)
all_dsps$dba <- gsub("111","1:11", all_dsps$dba)
all_dsps$dba <- gsub("ORGANICS","ORGANIC", all_dsps$dba)
all_dsps$dba <- gsub("ALPENGLOW BOTANICALS","ALPENGLOW", all_dsps$dba)
all_dsps$dba <- gsub("ALTITUDE ORGANIC CANNABIS","ALTITUDE ORGANICS", all_dsps$dba)
all_dsps$dba <- gsub("ALTITUDE ORGANIC MEDICINE","ALTITUDE ORGANICS", all_dsps$dba)
all_dsps$dba <- gsub("ANNIE?S DISPENSARY","ANNIES DISPENSARY", all_dsps$dba)
all_dsps$dba <- gsub("ASCEND CANNABIS.","ASCEND CANNABIS", all_dsps$dba)
all_dsps$dba <- gsub("BACK TO THE GARDEN HEALTH AND WELLNESS CENTER","BACK TO THE GARDEN", all_dsps$dba)
all_dsps$dba <- gsub("BACK TO THE GARDENS","BACK TO THE GARDEN", all_dsps$dba)
all_dsps$dba <- gsub("BEAN WELLNESS GROUP II","BEAN WELLNESS GROUP", all_dsps$dba)
all_dsps$dba <- gsub("BEST LORADO CANNABIS","BEST LORADO MEDS", all_dsps$dba)
all_dsps$dba <- gsub("BEST LORADO MEDS ORG","BEST LORADO MEDS", all_dsps$dba)
all_dsps$dba <- gsub("BEST LORADO MEDS.ORG","BEST LORADO MEDS", all_dsps$dba)
all_dsps$dba <- gsub("BESTLORADOMEDS.ORG","BEST LORADO MEDS", all_dsps$dba)
all_dsps$dba <- gsub("BESTLORADOMEDS.ORG","BEST LORADO MEDS", all_dsps$dba)
all_dsps$dba <- gsub("BONFIRE CANNABIS MPANY","BONFIRE CANNABIS", all_dsps$dba)
all_dsps$dba <- gsub("BOULDER MARIJUANA.","BOULDER MARIJUANA", all_dsps$dba)
all_dsps$dba <- gsub("BOULDER WELLNESS CANNABIS","BOULDER WELLNESS", all_dsps$dba)
all_dsps$dba <- gsub("BRECKENRIDGE ORGANIX","BRECKENRIDGE ORGANIC THERAPHY", all_dsps$dba)
all_dsps$dba <- gsub("BRECKENRIDGE CANNABIS CLUB","BRECKENRIDGE ORGANIC THERAPHY", all_dsps$dba)
all_dsps$dba <- gsub("CLEARANCE CANNABIS.","CLEARANCE CANNABIS", all_dsps$dba)
all_dsps$dba <- gsub("CRESTED BUTTE WELLNESS CENTER","CRESTED BUTTE", all_dsps$dba)
all_dsps$dba <- gsub("CRESTED BUTTE ALTERNTIVE MEDICINE","CRESTED BUTTE", all_dsps$dba)
all_dsps$dba <- gsub("DO CRESTED BUTTE","CRESTED BUTTE", all_dsps$dba)
all_dsps$dba <- gsub("DOCTOR?S ORDERS","DOCTORS ORDERS", all_dsps$dba)
all_dsps$dba <- gsub("ENLIGHTENED CARE PLUS","ENLIGHTENED CARE", all_dsps$dba)
all_dsps$dba <- gsub("FIFTY 2 EIGHTY","FIFTY2EIGHTY", all_dsps$dba)
all_dsps$dba <- gsub("GM ENTERPRISE.","GM ENTERPRISE", all_dsps$dba)
all_dsps$dba <- gsub("GREEN DREAM CANNABIS","GREEN DREAM", all_dsps$dba)
all_dsps$dba <- gsub("GREEN DREAM HEALTH SERVICES","GREEN DREAM", all_dsps$dba)
all_dsps$dba <- gsub("GREEN GRASS CENTRAL CITY","GREEN GRASS", all_dsps$dba)
all_dsps$dba <- gsub("GREEN FIELDS","GREENFIELDS", all_dsps$dba)
all_dsps$dba <- gsub("HEADQUARTERS CANNABIS","HEADQUARTERS", all_dsps$dba)
all_dsps$dba <- gsub("HEALING HOUSE DENVER","HEALING HOUSE", all_dsps$dba)
all_dsps$dba <- gsub("HEALING HOUSE LAKEWOOD","HEALING HOUSE", all_dsps$dba)
all_dsps$dba <- gsub("HELPING HANDS CANNABIS","HELPINGHANDS", all_dsps$dba)
all_dsps$dba <- gsub("HELPING HANDS HERBALS","HELPINGHANDS", all_dsps$dba)
all_dsps$dba <- gsub("HIGH COUNTRY HEALING 4","HIGH COUNTRY HEALING", all_dsps$dba)
all_dsps$dba <- gsub("HIGH COUNTRY HEALING 5","HIGH COUNTRY HEALING", all_dsps$dba)
all_dsps$dba <- gsub("HIGH COUNTRY HEALING 6","HIGH COUNTRY HEALING", all_dsps$dba)
all_dsps$dba <- gsub("HIGH COUNTRY HEALING 8","HIGH COUNTRY HEALING", all_dsps$dba)
all_dsps$dba <- gsub("IHC/SANTE","IHC/SANTE", all_dsps$dba)
all_dsps$dba <- gsub("IVITA WELLNESS","IVITA", all_dsps$dba)
all_dsps$dba <- gsub("KIND LOVE / KL","KIND LOVE", all_dsps$dba)
all_dsps$dba <- gsub("KL","KIND LOVE", all_dsps$dba)
all_dsps$dba <- gsub("KIND LOVE EXPRESS","KIND LOVE", all_dsps$dba)
all_dsps$dba <- gsub("KIND MEDS","KINDMEDS", all_dsps$dba)
all_dsps$dba <- gsub("LA NTES LNE BAR + DISPENSARY","LANTES CLONE BAR AND DISPENSARY", all_dsps$dba)
all_dsps$dba <- gsub("LANTES CLONE BAR + DISPENSARY","LANTES CLONE BAR AND DISPENSARY", all_dsps$dba)
all_dsps$dba <- gsub("LEAF ASPEN","LEAF", all_dsps$dba)
all_dsps$dba <- gsub("LEVELS IV.","LEVELS IV", all_dsps$dba)
all_dsps$dba <- gsub("LIGHTSHADE LABS","LIGHTSHADE", all_dsps$dba)
all_dsps$dba <- gsub("LIVWELL III","LIVWELL", all_dsps$dba)
all_dsps$dba <- gsub("LORADO HARVEST SUPPLY","LORADO HARVEST", all_dsps$dba)
all_dsps$dba <- gsub("LORADO HARVEST COMPANY","LORADO HARVEST", all_dsps$dba)
all_dsps$dba <- gsub("LOTUS MEDICAL II","LOTUS MEDICAL", all_dsps$dba)
all_dsps$dba <- gsub("MAGGIE?S FARM","MAGGIES FARM", all_dsps$dba)
all_dsps$dba <- gsub("MAGNOLIA ROAD CANNABIS COMPANY","MAGNOLIA ROAD", all_dsps$dba)
all_dsps$dba <- gsub("MAGNOLIA ROAD CANNABIS","MAGNOLIA ROAD", all_dsps$dba)
all_dsps$dba <- gsub("MAGNOLIA ROAD CANNABIS","MAGNOLIA ROAD", all_dsps$dba)
# removes all of the trailing names conventions
all_dsps$dba <- gsub("MEDICINE MAN(?!.)$","MEDICINE MAN", all_dsps$dba, perl = TRUE )
all_dsps$dba <- gsub("MMJ AMERICA(?!.$)","MMJ AMERICA", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("MOUNTAIN MEDICINALS(?!.)$","MOUNTAIN MEDICINALS", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("COMPASSIONATE CARE(?!.)$","COMPASSIONATE CARE", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("MR. NICE GUYS","MR NICE GUYS", all_dsps$dba) #
all_dsps$dba <- gsub("NATIVE ROOTS(?!.)$","NATIVE ROOTS", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("NATURES HERBS(?!.)$","NATURES HERBS", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("NATURES MEDICINE(?!.)$","NATURES MEDICINE", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("NEW AMSTERDAM(?!.)$","NEW AMSTERDAM", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("NORTHERN LIGHTS CANNABIS.","NORTHERN LIGHTS CANNABIS", all_dsps$dba)
all_dsps$dba <- gsub("PATIENTS CHOICE(?!.)$","PATIENTS CHOICE", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("PEAK","PEAK DISPENSARY", all_dsps$dba)
all_dsps$dba <- gsub("PINK HOUSE(?!.)$","PINK HOUSE", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("PREFERRED ORGANIC(?!.)$","PREFERRED ORGANIC", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("PRIMO","PRIMO CANNABIS", all_dsps$dba)
all_dsps$dba <- gsub("PURE MARIJUANA DISPENSARY","PURE DISPENSARY", all_dsps$dba) #
all_dsps$dba <- gsub("PURE MEDICAL DISPENSARY","PURE DISPENSARY", all_dsps$dba)
all_dsps$dba <- gsub("PURE MEDICAL","PURE DISPENSARY", all_dsps$dba)
all_dsps$dba <- gsub("RESERVE 1(?!.)$","RESERVE 1", all_dsps$dba, perl = TRUE) #
all_dsps$dba <- gsub("RIVERROCK(?!.)$","RIVERROCK", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("ROOTS RX","ROOTSRX", all_dsps$dba)
all_dsps$dba <- gsub("SMOKEY?S","SMOKEYS", all_dsps$dba)
all_dsps$dba <- gsub("SMOKED(?!.)$","SMOKED", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("SMOKING GUN","SMOKINGUN APOTHECARY", all_dsps$dba)
all_dsps$dba <- gsub("STAR BUDS","STARBUDS", all_dsps$dba)
all_dsps$dba <- gsub("STEEL CITY MEDS(?!.)$","STEEL CITY MEDS", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("STICKY BUDS(?!.)$","STICKYBUDS", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("STICKY BUDS","STICKYBUDS", all_dsps$dba)
all_dsps$dba <- gsub("TETRA HYDRO CENTER","TETRAHYDROCENTER", all_dsps$dba)
all_dsps$dba <- gsub("THE CANARY?S SONG","THE CANARYS SONG", all_dsps$dba)
all_dsps$dba <- gsub("THE CLINIC(?!.)$","THE CLINIC", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("THE MEDICINE MAN","MEDICINE MAN", all_dsps$dba)
all_dsps$dba <- gsub("THE SANCTUARY(?!.)$","THE SANCTUARY", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("THE STONE","THE STONE DISPENSARY", all_dsps$dba)
all_dsps$dba <- gsub("TRU CANNABIS(?!.)$","TRU CANNABIS", all_dsps$dba, perl = TRUE)
all_dsps$dba <- gsub("TUMBLEWEEK DISPENSARY EDWARDS","TUMBLEWEED DISPENSARY EDWARDS", all_dsps$dba)
all_dsps$dba <- gsub("UNITY RD. CANNABIS SHOP","UNITY ROAD", all_dsps$dba)
all_dsps$dba <- gsub("WALKING RAVEN II","WALKING RAVEN", all_dsps$dba)
all_dsps$dba <- gsub("WOLF PAC CANNABIS","WOLFPAC CANNABIS", all_dsps$dba)
all_dsps$dba <- gsub("ALLGREENS","CANNABIS MEDICAL TECHNOLOGY", all_dsps$dba)
all_dsps$dba <- gsub("3D CANNABIS CENTER AND 3D DISPENSARY","3D CANNABIS CENTER", all_dsps$dba)This process takes a few minutes, so I exported the data for manual review.
Code
write.csv(all_dsps,
"~/gvsu/winter 23/CIS 635/NIMBY/cleaning/counting_dispensaries/dps_data_cleaned/dispensaries.csv")
identify and label dispensaries by their type
There are several type of licenses recorded by the Colorado Department of Revenue. NIMBY focuses on recreational and medical dispensaries, so those are the businesses I have identified based on the license number as shown below.
Code
cleaned_dsps <- all_dsps
cleaned_dsps <- cleaned_dsps %>%
mutate(type = if_else(startsWith(license_no, "402R"), "retail",
if_else(startsWith(license_no, "402-"), "medical",
"other")))
#remove the testing facilities, growers and transporters
dsps <- cleaned_dsps %>%
filter(type == "medical" | type == "retail")
dsps$dba <- ifelse(dsps$dba == "NONE", NA, dsps$dba) # change the None's to NAs
dsps$zip<- substr(dsps$zip, start = 1, stop = 5) # remove trailing numbers
find missing business names
adapted from “Dplyr solution using slice and group”
Along with variations in spelling, some years the “doing business as” field was left blank. Assuming the license number is only assigned to the same business identified in another time period, I can use that information to create a master list of business names to match against.
Code
# select all business names and license numbers
dsps_list <-dsps %>%
select(dba, license_no) #
# remove the missing fields, coming for those later
dsps_list <- as.data.frame(na.omit(dsps_list)) # remove na
# keep only the first instance (since their are multiple naming conventions
# for each licensee)
dsps_list <- dsps_list %>%
group_by(license_no) %>%
slice(1)
# merge the business names by the license number
# all.x = TRUE argument means include all rows from dsps
# even if there is no matching row in dsps_list
dsps_merge <- merge(dsps, dsps_list, by = "license_no", all.x= TRUE)
#update the "doing business as" column, replace the N/As
dsps_merge$dba <- coalesce(dsps_merge$dba.y, dsps_merge$dba.x)
# if that still doesn't work, update the umatched licence numbers with the licensee name
dsps_merge$dba <- coalesce(dsps_merge$dba, dsps_merge$licensee)
# remove the duplicated columns
dsps_merge <- dsps_merge %>%
select(
license_no,
dba,
city,
zip,
type,
date,
year)Here is the new cleaned list of businesses with limited null fields!
Code
head(dsps_merge) license_no dba city zip type date year
1 402-00001 CANNABIS MEDICAL TECHNOLOGY Denver 80204 medical 2020-08-01 2020
2 402-00001 CANNABIS MEDICAL TECHNOLOGY Denver 80204 medical 2016-02-01 2016
3 402-00001 CANNABIS MEDICAL TECHNOLOGY Denver 80204 medical 2020-12-01 2020
4 402-00001 CANNABIS MEDICAL TECHNOLOGY Denver 80204 medical 2015-04-01 2015
5 402-00001 CANNABIS MEDICAL TECHNOLOGY Denver 80204 medical 2013-11-01 2013
6 402-00001 CANNABIS MEDICAL TECHNOLOGY Denver 80204 medical 2016-05-01 2016
find the missing zip codes
This same issue occurs with the zip code field, I apply the same technique used to find missing businesses.
Code
dsps_zips <-dsps_merge %>%
select(zip, city, license_no) # add type for a distinct list
dsps_zips <- as.data.frame(na.omit(dsps_zips)) # remove na
# keep only the main/ first instance
dsps_zips<- dsps_zips %>%
group_by(license_no) %>%
slice(1)
#save this for creating per capita variables
write.csv(dsps_zips,
"~/gvsu/winter 23/CIS 635/NIMBY/cleaning/counting_dispensaries/dps_data_cleaned/dsps_zips.csv")
# xlook-up R style: overide the na's with the matches, same as a left_join
dsps_merge_zips <- merge(dsps_merge, dsps_zips, by = "license_no", all.x= TRUE)
dsps_merge_zips$zip <- coalesce(dsps_merge_zips$zip.x, dsps_merge_zips$zip.y)
# remove the duplicated columns, and update the main data frame
storelist<- dsps_merge_zips %>%
select(
license_no,
dba,
type,
city = city.x,
zip,
date,
year
)
# export for manual review if needed
write.csv(storelist,
"~/gvsu/winter 23/CIS 635/NIMBY/cleaning/counting_dispensaries/dps_data_cleaned/dispensaries.csv")
find dispensaries that are both medical and recreational sellers
adapted from “Join Data with dplyr in R: 6 Examples”
This is where my methodology differs drastically from the original paper. NIMBY matches the physical locations to determine which licensees are both medical and recreational (or retail).
Our algorithm utilizes a customized cross-walk that identifies licenses assigned to the same address (e.g. matches 250 W. Colfax to 250 West Colfax) and identifies the co-location of medical and recreational based on the name of the business holding the licenses (since recreational licensees were granted only to the license holders of medical license).
[@brinkman2017]
My approach to creating three buckets of dispensaries:
divide the data into two sets; “medical” and “retail” ,
cross compare the lists, and identify where a business name appears in the “both” categories during the same time period
remove the collocations from the retail and medical businesses
Code
# divide this into two new dataframes, one for medical, one for recreational
med_dsp <- filter(storelist, type == "medical")
rec_dsp <- filter(storelist, type == "retail")
# find the business that have the same name, city and date
# with two different names, so this is my best estimate
both_dsp <- merge(med_dsp, rec_dsp, by = c("dba", "date"))%>%
select(
dba,
"city" = `city.x`,
date,
"year" = `year.x`,
"zip" = `zip.x`)
both_dsp$type <- "both"
# remove duplicates
both_dsp <- both_dsp %>% distinct(dba, date, type, .keep_all = TRUE)
# removes the "both" businesses from the respective dataframes
med_only <- med_dsp %>%
anti_join(both_dsp, by = c("dba", "date"))
med_only$type <- "med_only" # label the type
rec_only <- rec_dsp %>%
anti_join(both_dsp, by = c("dba", "date"))
rec_only$type <- "rec_only" # label the type
# combine the dataframes together (like if i were to copy and paste it at the bottom of an excel file)
storelist_cleaned <- bind_rows(med_only,both_dsp, rec_only )
storelist_cleaned <- storelist_cleaned %>% distinct(dba, type, date, zip, .keep_all = TRUE)
write.csv(storelist_cleaned,
"dispensary_types_defined.csv")Graph the Results for Analysis:
Code
# Summarize the results
# dsps_growth$date <- ymd(dsps$date)
# dsps_growth$type <- factor(dsps_growth$type,
# levels=c("med_only", "both", "rec_only"))
# dsps_growth$type <- factor(dsps_growth$type, levels = rev(c("med_only", "both", "rec_only")))
dsps_growth <- storelist_cleaned %>%
filter( year < 2017) %>%
select(type,
dba,
date) %>%
group_by(date, type) %>%
summarise(num_licenses = n())`summarise()` has grouped output by 'date'. You can override using the
`.groups` argument.Code
# create a graph
fig1.b <- ggplot(dsps_growth, aes(x = date, y = num_licenses,
fill = factor(type))) +
geom_area(alpha=0.6 , size=.5, colour="white")+
scale_fill_manual(values =c("#B41876","#23CB7F", "#1679A6")) +
ylim(0, 800) +
geom_vline(xintercept = as.Date("2014-01-01"), linetype = "dashed") +
geom_text(x = as.Date("2014-09-01") + 30,
y = 750, label = "first recreational sales",
family = "Times New Roman") +
annotate("text", x = as.Date("2015-01-01"), y = 500, label = "both") +
annotate("text", x = as.Date("2015-01-01"), y = 300, label = "medical only") +
annotate("text", x = as.Date("2015-01-01"), y = 50, label = "rec only") +
labs(y="Number of Stores",
x ="Year",
color = "Type",
title = "Dispensary Growth",
subtitle = "State of Colorado 2013–2016",
hjust = 0, size = 5) +
theme_minimal() +
theme(legend.position = "none")Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.Code
fig1.bWarning in grid.Call.graphics(C_text, as.graphicsAnnot(x$label), x$x, x$y, :
font family not found in Windows font database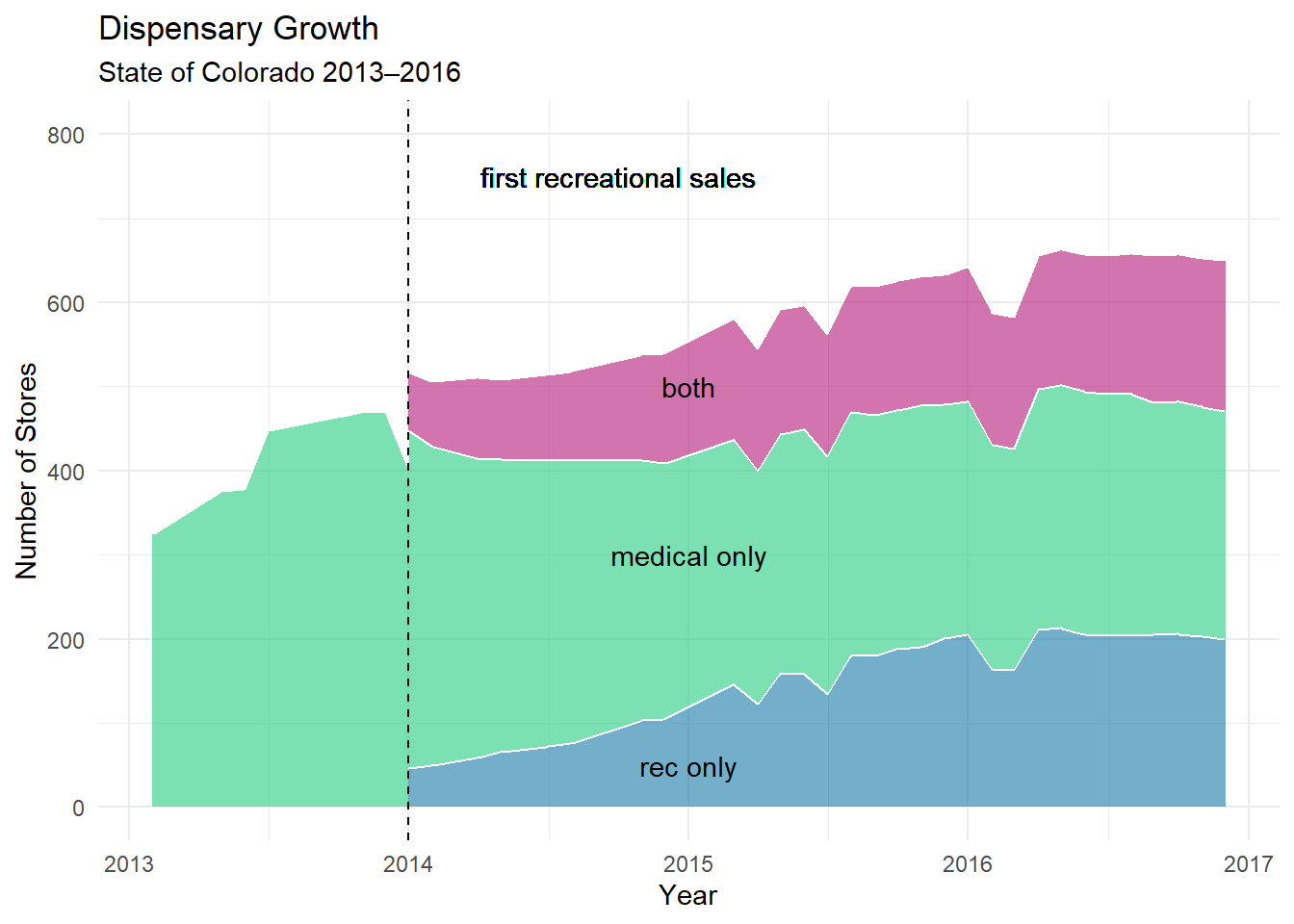
Code
ggsave("images/dsp_growth.png")Saving 7 x 5 in imageWarning in grid.Call.graphics(C_text, as.graphicsAnnot(x$label), x$x, x$y, :
font family not found in Windows font database